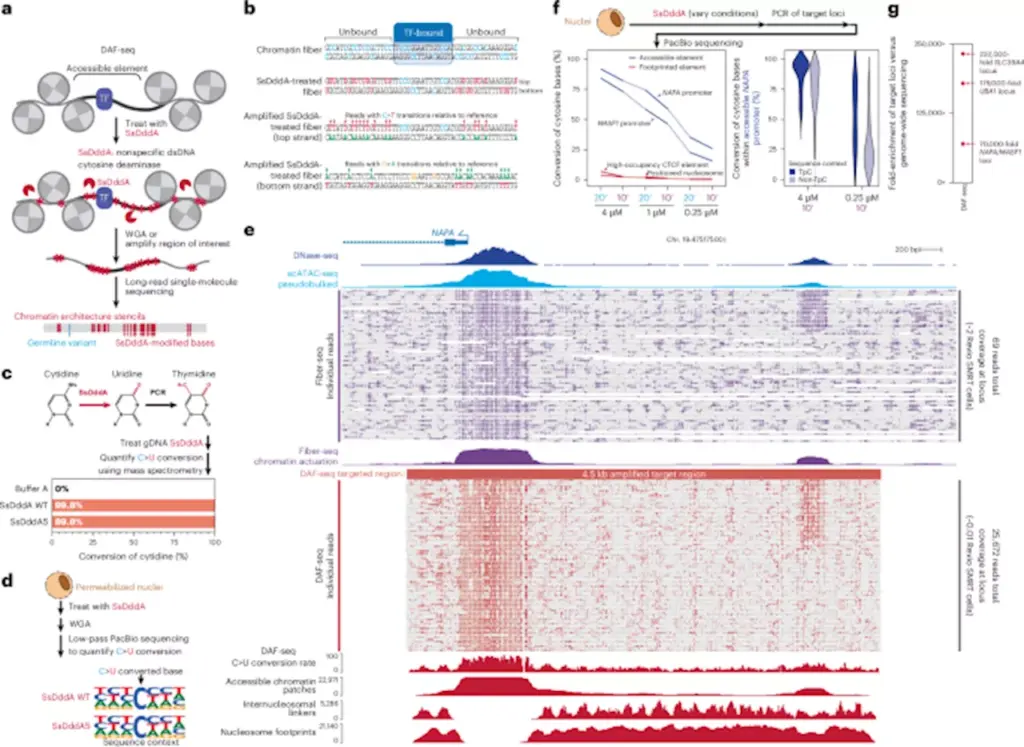
A team of researchers has introduced a revolutionary technique called Deaminase-Assisted single-molecule chromatin Fiber sequencing (DAF-seq), which offers unprecedented insights into gene regulation by mapping chromatin fiber architectures at a single-cell level. This innovative approach allows scientists to observe cooperative protein occupancy across chromosome-length fibers, providing a clearer understanding of how genetic variation and chromatin states influence regulatory mechanisms within diploid organisms.
DAF-seq, developed by researchers at the University of Washington and Washington University in St. Louis, addresses the challenges posed by heterogeneity in gene occupancy between haplotypes and cells. By enabling single-molecule footprinting with near-nucleotide resolution, DAF-seq synchronously profiles chromatin states and DNA sequences, revealing the intricate dynamics of gene regulation.
The implementation of single-cell DAF-seq has resulted in comprehensive chromosome-length protein co-occupancy maps that cover 99% of each cell’s mappable genome. Findings indicate significant chromatin plasticity within and between individual diploid cells, with chromatin actuation diverging by 61% between haplotypes within a single cell and 63% between different cells. This substantial variation underscores the complexity of chromatin interactions and their functional consequences.
Additionally, the research highlights that regulatory elements tend to co-actuate along the same chromatin fiber. This behavior is influenced by a distance-dependent mechanism akin to cohesin-mediated loops, which further complicates our understanding of regulatory networks. The detailed maps generated by DAF-seq not only illuminate the cooperative interactions among proteins but also reveal the functional impact of somatic variants and rare chromatin epialleles.
The study, led by A.B. Stergachis and E.G. Stergachis, has garnered substantial support from various funding bodies, including the Burroughs Wellcome Fund and the Chan Zuckerberg Initiative. The research was facilitated by the National Institutes of Health Common Fund, under the award number UM1DA058220. This collaborative effort involved contributions from multiple institutions, emphasizing the importance of interdisciplinary research in advancing our understanding of genetics.
As the implications of DAF-seq unfold, the potential to characterize protein occupancy across entire chromosomes with single-nucleotide, single-molecule, single-haplotype, and single-cell precision could pave the way for significant advancements in genetics and molecular biology. The approach promises to enhance our understanding of complex genetic interactions and may lead to novel insights into the regulation of gene expression.
The full findings from this research are detailed in a manuscript available on the Stergachis Lab GitHub page. Through this significant advancement in chromatin mapping technology, researchers are poised to explore new avenues in genomic research, ultimately contributing to the broader understanding of human genetics and disease.






