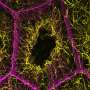
Researchers have successfully created a comprehensive three-dimensional map of human chromosomes, detailing over 140,000 DNA loops within the nucleus of human cells. This groundbreaking work, published in the scientific journal Nature, provides an unprecedented look at the complex organization and folding of chromosomes, which is essential for understanding cellular function and genetic regulation.
The study, led by a collaborative team from Stanford University and the University of California, San Francisco (UCSF), reveals how DNA is structured and interacts within the nucleus. This mapping effort not only enhances our knowledge of DNA architecture but also opens new avenues for research into genetic diseases and developmental biology.
Insights into Chromosomal Organization
The research team utilized advanced imaging techniques to visualize the intricate arrangement of chromosomes. By analyzing the spatial relationships among DNA loops, the scientists uncovered how these loops form distinct domains within the nucleus. These domains play a crucial role in gene expression and cellular processes.
The data indicate that the organization of chromosomes is far more complex than previously understood. Each loop connects different regions of the genome, facilitating communication between genes that may be located far apart on the linear DNA strand. This intricate network of interactions is vital for orchestrating the proper functioning of genes, influencing everything from cell differentiation to responses to environmental stimuli.
Implications for Genetic Research
The implications of this research extend beyond basic biology. Understanding the architecture of human chromosomes can significantly impact the study of genetic disorders. Many diseases arise from disruptions in genomic organization, and this detailed map may guide future investigations into the mechanisms behind these disorders.
For instance, the findings could aid in pinpointing the genetic basis of certain cancers and developmental conditions. With more precise knowledge of chromosomal organization, researchers can develop targeted therapies that address the root causes of these diseases.
In addition, this work sets the stage for further explorations into epigenetics, the study of how environmental factors influence gene expression without altering the underlying DNA sequence. The research team anticipates that their findings will spur additional studies aimed at unraveling the complexities of gene regulation and its implications for health and disease.
As scientists continue to delve deeper into the world of genetics, the mapping of DNA loops provides a crucial building block for future discoveries. The advancements made in this study not only enhance our understanding of human biology but also pave the way for innovative approaches to medical research and treatment.